Return to DNA Test Results Index
Link to table where Y chromosome marker results for all participants are summarized together
Y Chromosome Test Results For Harvey Settler
Descendant G. Alan Robison
Y-Chromosome test results for Al Robison using 25 markers. Family Tree DNA interprets this haplotype as belonging to the Haplogroup I1A (Norse) lineage prominent amongst Norwegian Vikings and quite common amongst Danish Vikings who invaded and settled in North Eastern England in the 10th century. Al is a direct male descendant of William Robison (6 Sep 1811 - 16 Apr 1889), one of the original 1837 settlers of Harvey, NB who was a native of the Borders region, as well as William's father Marshall Robison I (1787 - 11 Apr 1862) who came to Harvey settlement in 1842.
The test results below are from the laboratory analysis of Al's Y-chromosome. His DNA was analyzed for Short Tandem Repeats (STRs), which are repeating segments of the genome that have a high mutation rate. See definitions of Locus, DYS#, and Alleles at the bottom of this page.
Locus
DYS#
Alleles
1
393
13
2
390
23
3
19 (also 394)
14
4
391
10
5
385a
14
6
385b
14
7
426
11
8
388
14
9
439
11
10
389-1
13
11
392
11
12
389-2
29
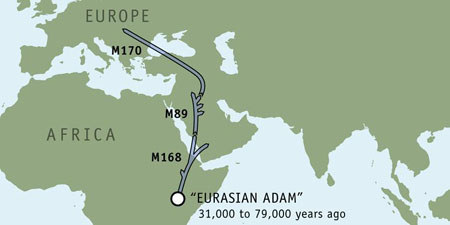
13 458 14 14 459a 8 15 459b 9 16 455 8 17 454 11 18 447 23 19 437 16 20 448 20 21 449 28 22 464a 12 23 464b 13 24 464c 14 25 464d 15 Through use of a haplogroup predictor algorithm developed by Whit Athey, Al's Y chromosome results identify him as a member of haplogroup I1a, a lineage defined by a genetic marker called M170. This haplogroup is the final destination of a genetic journey that began some 60,000 years ago with an ancient Y chromosome marker called M168.
The very widely dispersed M168 marker can be traced to a single individual—"Eurasian Adam." This African man, who lived some 31,000 to 79,000 years ago, is the common ancestor of every non-African person living today. His descendants migrated out of Africa and became the only lineage to survive away from humanity's home continent.
Population growth during the Upper Paleolithic era may have spurred the M168 lineage to seek new hunting grounds for the plains animals crucial to their survival. A period of moist and favorable climate had expanded the ranges of such animals at this time, so these nomadic peoples may have simply followed their food source.
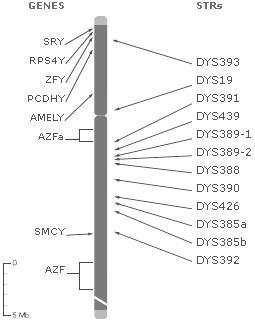
The location on the Y chromosome of each of the tested markers is depicted in the image above, with the number of repeats for each of your STRs presented to the right of the marker. For example, DYS19 is a repeat of TAGA, so if Dallas's DNA repeated that sequence 14 times at that location, it would appear: DYS19 = 14. Studying the combination of these STR lengths in a persons Y Chromosome allows researchers to place individuals in a haplogroup, which reveals the complex migratory journeys of ancestors.
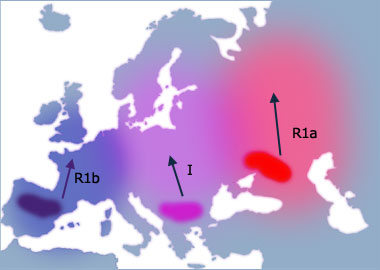
By 12,000 Y BP the ice has retreated and the land became much more supportive to life. Many animal species returned to inhabit the land. The three groups of humans had taken refuge for so long that their DNA had naturally picked up mutations, and consequently can be defined into different haplogroups. As they spread from these refuges, Haplogroups R1b, I and R1a propagated across Europe. The three groups of humans had taken refuge for so long that their DNA had naturally picked up mutations, and consequently can be defined into different haplogroups. As they spread from these refuges, Haplogroups R1b, I and R1a propagated across Europe. Haplogroup R1b is common on the western Atlantic coast as far as Scotland. Haplogroup I is common across central Europe and up into Scandinavia. Haplogroup R1a is common in eastern Europe and has also spread across into central Asia and as far as India and Pakistan.
Improved tools and rudimentary art appeared during this same epoch, suggesting significant mental and behavioral changes. These shifts may have been spurred by a genetic mutation that gave "Eurasian Adam's" descendants a cognitive advantage over other contemporary, but now extinct, human lineages.
Some 90 to 95 percent of all non-Africans are descendants of the second great human migration out of Africa, which is defined by the marker M89.
M89 first appeared 45,000 years ago in Northern Africa or the Middle East. It arose on the original lineage (M168) of "Eurasian Adam," and defines a large inland migration of hunters who followed expanding grasslands and plentiful game to the Middle East.
Many people of this lineage remained in the Middle East, but others continued their movement and followed the grasslands through Iran to the vast steppes of Central Asia. Herds of buffalo, antelope, woolly mammoths, and other game probably enticed them to explore new grasslands.
With much of Earth's water frozen in massive ice sheets, the era's vast steppes stretched from eastern France to Korea. The grassland hunters of the M89 lineage traveled both east and west along this steppe "superhighway" and eventually peopled much of the continent.
A group of M89 descendants moved north from the Middle East to Anatolia and the Balkans, trading familiar grasslands for forests and high country. Though their numbers were likely small, genetic traces of their journey are still found today.
Haplogroup I, is widespread throughout southeastern and central Europe and most common in the Balkans. Members of this haplogroup carry a 20,000-year-old marker dubbed M170.
This Y chromosome marker first appeared in the Middle East. Its subsequent spread into southeastern Europe may have accompanied the expansion of the prosperous Gravettian culture. These Upper Paleolithic people used effective communal hunting techniques and developed art notable for voluptuous female carvings often dubbed "Venus" figures.
The later spread of this lineage could be also tied to the mid-first millennium B.C. Celtic culture. The tantalizing possibility could explain the wider dispersal of this unique genetic marker.
Interpretation of the various population varieties within Y-Haplogroup I and their extended Modal Haplotypes has been studied in some detail by has been carried out by Ken Nordtvedt and is summarized on his web site.
Al's haplotype is typical of I1a in having a value of DYS455 = 8, as almost all members of this haplogroup have the very unusual 8 repeats at DYS455, a very slow mutating marker. Virtually no other European haplotypes outside of I1a have 8 repeats at DYS455. This makes identifying and studying I1a haplotypes quite straightforward. The motif YCAIIa,b = 19,21 is also close to universal over all of I1a, however I1c shares this same modal pair of repeat values at this marker, so one would look first to DYS455 before YCAIIa,b in identifying I1a haplotypes.
Almost all I1a has the very unusual 8 repeats at DYS455, a very slow mutating marker. And virtually no other European haplotypes outside of I1a have 8 repeats at DYS455. This makes identifying and studying I1a haplotypes quite straightforward if one's extended haplotypes include this marker. The motif YCAIIa,b = 19,21 is also close to universal over all of I1a, although I1c shares this same modal pair of repeat values at this marker, so one would look first to DYS455 before YCAIIa,b in identifying I1a haplotypes.
Al is part of a very distinctive subgroup of haplogroup I1a termedI1a-N (Norse). This is the most populous form of I1a found in Sweden and Finland, and is a close second in Norway. It is found in only tiny quantities in continental Europe south of the Baltic and North Seas, but takes second place to I1a-AS in Denmark. With its shifts at DYS390 --- 22 to 23 --- and at DYS385a,b --- 13,14 to 14,14 --- the core Norse haplotype to look for is:
DYS19 = 14
DYS390 = 23
DYS385a,b = (14,14)
DYS391 = 10
DYS 392 = 11
DYS 393 = 13
DYS 389i,ii = (12,28)
DYS 388 = 1414,23,(14,14),10,11,13,(12,28),14 at the classical markers listed previously. But the strongly confirmative shift for identifying a Norse I1a rather than Anglo-Saxon I1a is at DYS462 where there should now be 13 repeats. Also supporting the distinct Norse variety is the strong dominance 12,14,15,16 at DYS464a,b,c,d (41 to 5) over the 12,14,15,15 motif at this 4-copy marker.
Definitions
Allele: Alternative form of a genetic locus; a single allele for each locus is inherited separately from each parent (e.g., at a locus for eye color the allele might result in blue or brown eyes).
DYS : D = DNA, Y = Y chromosome, S = a unique DNA segment. This nomenclature is controlled by the HUGO Gene Nomenclature Committee, with the assignment of new DYS numbers. This guideline determines each part of the symbol for naming arbitrary DNA fragments and loci. See section Appendix App 1.1 DNA Segments located at:
http://www.gene.ucl.ac.uk/nomenclature/guidelines.html#1.4Locus (pl. loci): The position on a chromosome of a gene or other chromosome marker; also, the DNA at that position. The use of locus is sometimes restricted to mean regions of DNA that are expressed. The specific physical location of a gene on a chromosome. From the Latin for 'place'. A stretch of DNA at a particular place on a particular chromosome — often used for a 'gene' in the broad sense, meaning a stretch of DNA being analyzed for variability (e.g., a microsatellite locus).
Marker: An identifiable physical location on a chromosome (e.g., restriction enzyme cutting site, gene) whose inheritance can be monitored. Markers can be expressed regions of DNA (genes) or some segment of DNA with no known coding function but whose pattern of inheritance can be determined. A gene of known location on a chromosome and phenotype that is used as a point of reference in the mapping of other loci.
Return to DNA Test Results Index
Link to table where Y chromosome marker results for all participants are summarized together