Return to DNA Test Results Index
Link to table where Y chromosome marker results for all participants are summarized together.
Migration History of the Early Ancestors of Tim Patterson and Sons Malcolm and Calder
Based on Y Chromosome Test Results
(Haplogroup R1b1)
In August 2005 a 12 marker Y-chromosome test results for R. Tim Patterson came back from the National Geographic Society's Genographic Project, which he subsequently upgraded to to 37 markers with Family Tree DNA in October, 2005. Family Tree DNA interprets this haplotype as belonging to Haplogroup R1b (M343), the most common lineage in Western Europe. In late fall 2005 samples for Tim and his sons Malcolm and Calder were also submitted to the Sorenson Molecular Genealogy Foundation (SMGF) for testing. All markers tested in common by the two laboratories returned identical results. An 11 additional markers were obtained by the SMGF testing bringing the total markers tested for Tim to 48. The Pattersons are direct male descendant of William James Patterson (6 Mar 1812 - 8 Dec 1892) who according to his obituary was born in Berwick Upon Tweed, in the Borders region of Northumberland, United Kingdom.
The test results below are from the laboratory analysis of Tim, Malcolm and Calder's Y-chromosome. Their DNA was analyzed for Short Tandem Repeats (STRs), which are repeating segments of the genome that have a high mutation rate. See definitions of Locus, DYS#, and Alleles at the bottom of this page.
Earliest Known
Male Ancestor
William Patterson*
b 6 Mar 1812
Berwick, UK
d 12 Dec 1892 Harvey, NBWilliam Patterson*
b 6 Mar 1812
Berwick, UK
d 12 Dec 1892 Harvey, NBWilliam Patterson*
b 6 Mar 1812
Berwick, UK
d 12 Dec 1892 Harvey, NBTested Participant R. Tim Patterson Malcolm A.
PattersonCalder W.
PattersonKit FTDNA N7352
& Sorenson
Sorenson Sorenson Haplogroup R1b1c R1b1c R1b1c SNP Locus DYS # Alleles Alleles Alleles Panel 1 1
393
14
14 14 2
390
25
25 25 3
19 (also 394)
14
14 14 4
391
11
11 11 5
385a
11
11 11 6
385b
14
14 14 7
426
12
12 12 8
388
12
12 12 9
439
11 11 11 10
389-1
13
13 13 11
392
14
14 14 12
389-2
29
29 29 Panel 2 13
458
18 18 18 14
459a
9 9 9 15
459b
9 9 9 16
455
11 11 11 17
454
11 11 11 18
447
25 25 25 19
437
15 15 15 20
448
19 19 19 21
449
29 29 29 22
464a**
15 15 15 23
464b**
15 15 15 24
464c**
17 17 17 25
464d**
18 18 18 Panel 3 26
460
11
11 11 27
GATA H4
11
11 11 28
YCA II a
19
19 19 29
YCA II b
23
23 23 30
456
17
17 17 31
607
14
32
576
17
33
570
17
34
CDY a
37
35
CDY b
40
36
442
12
12 12 37
438
12
12 12 Panel 4 38 461 12 12 12 39 462 11 11 11 40 YGATAA10 12 12 12 41 635/C4 24 24 24 42 GGAAT1B07 10 10 10 43 441 13 13 13 44 444 12 12 12 45 445 46 446 13 13 13 47 452 11 11 11 48 463 22 22 22 49 643 50 425 Panel 5 51 450 52 485 53 495 54 505 55 508 56 520 57 522 68 532 69 533 70 534 71 540 72 556 73 557 74 594
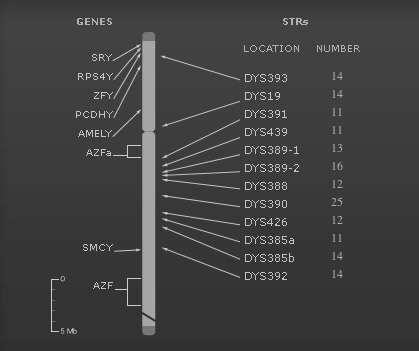
The location on the Y chromosome of each of the tested markers is depicted in the image, with the number of repeats for each of your STRs presented to the right of the marker. For example, DYS19 is a repeat of TAGA, so if Tim, Malcolm and Calder's DNA repeated that sequence 14 times at that location, it would appear: DYS19 14. Studying the combination of these STR lengths in a persons Y Chromosome allows researchers to place individuals in a haplogroup, which reveals the complex migratory journeys of ancestors.
The five year Genographic Project, which began in 2005, is sponsored by the National Geographic Society and the Waitt Institute for Historical Discovery, in cooperation with IBM. The project is dedicated to tracing human roots in deep time to a single origin. The goals of this ambitious global project are twofold: to capture a snapshot of human history locked within our DNA before it disappears forever, and to highlight the untold stories and uncertain future of indigenous peoples worldwide.
With funding from the Waitt Family Foundation and support from the Waitt Institute for Historical Discovery, scientists will establish 10 centers around the world and will study more than 100,000 DNA samples from indigenous populations. The project is expected to reveal rich details about global human migratory history and to drive new understanding about the connections and differences that make up the human species.
Genetic research has answered the origins question resoundingly-- we all ultimately trace back to Africa within the past 60,000 years. But we still need to discover many of the details of this journey. The Genographic Project will be able to map more points along this 'epic migration'.
Tim took part in the public part of the project by purchasing a Participation Kit and submitting his own cheek swab samples, allowing him to track the overall progress of the project as well as learn more about his own ancestral migratory history.
The resulting public database will house one of the largest collections of human population genetic information ever assembled and will serve as an unprecedented resource for geneticists, historians and anthropologists.
The Sorenson Molecular Genealogy Foundation is a non-profit organization dedicated to building the world's foremost collection of DNA and corresponding genealogical information.
SMGF is making their collection available for searching on this web site. Finding matching DNA results and pedigrees in the Sorenson Database can help you make new family connections throughout the world and across generations.
The Foundation is a world leader in DNA research with direct application to genealogy. Their work complements other studies that focus on the "deep ancestry" of humankind.
SMGF was inspired by discussions in 1999 between philanthropist James LeVoy Sorenson and BYU Professor Scott Woodward about using DNA in genealogy.
Since that time, SMGF has collected more than 60,000 DNA samples, including those of the Patterson's together with four-generation pedigree charts, from volunteers in more than 100 countries around the world.
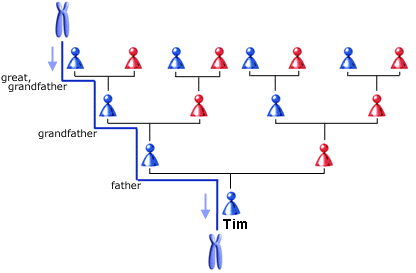
Based on analysis of his Y-chromosome Tim, Malcolm and Calder are members of Haplogroup R1b1 (M343), very common today throughout western Europe and North America. Since Y chromosomes mutate very slowly, and are directly passed down from father to son his Y chromosome is identical to that of his father, brother, paternal grandfather, children and so on back through the generations. This is why the results for Tim, and sons Malcolm and Calder are identical. In fact their Y-chromosomes are nearly identical to that of Tim's great great grandfather, William James Patterson (6 Mar 1812 - 8 Dec 1892) who emigrated to Canada in 1837 with the other settlers of Harvey Settlement aboard the Cornelius of Sunderland. All direct male descendants of William share basically the same Y chromosome, and there are undoubtedly many people in the borders area of the United Kingdom with the same Y chromosome; male descendants of Williams direct male ancestors.
We inherit a Y-chromosome from our father, who in turn received his from his father and so on. In fact, it can remain unchanged for many generations, so you will likely have the exact same Y-chromosome as your g-g-g-g-g-g-grandfather. You will also often share the same surname as it is passed down the very same line (which is why it is so useful to genealogists!)
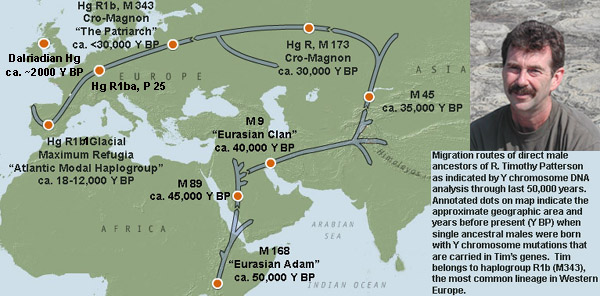
Over time Y-chromosomes do mutate though and in his DNA Tim can carries the lineage of the ancestral Y-chromosome M168 marker, which originated in Africa about 50,000 years before present (BP). This DNA marker is linked to a single individual "Eurasian Adam" who lived just north of lake Uganda, and who is the male ancestor of all Non-African people alive today.
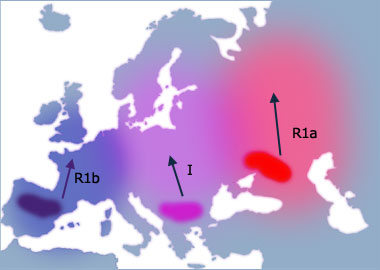
Migration routes of direct male ancestors of Tim, Malcolm and Calder Patterson as indicated by Y chromosome DNA analysis through last 50,000 years. Annotated dots on map indicate the approximate geographic area and years before present (Y BP) when ancestral males were born with Y chromosome mutations that are carried in Tim's genes. Tim belongs to haplogroup (Hg) R1b1, a subset of R1b, the most common lineage in Western Europe.
Eurasian Adam's M168 haplotype carrying descendents migrated from Africa about 50,000 years ago and became the only lineage to survive away from humanity's home continent. Population growth during the Upper Paleolithic era may have spurred the M168 lineage to seek new hunting grounds for the plains animals crucial to their survival. A period of moist and favorable climate had expanded the ranges of such animals at this time. Some nomadic peoples bearing the M168 marker left Africa as they followed their food source toward the Middle East. Improved tools and rudimentary art have been dated from this epoch, indicating significant mental and behavioral changes. These shifts may have been spurred by a genetic mutation that gave "Eurasian Adam's" descendants a cognitive advantage over other contemporary but now extinct, human lineages.
Their journey out of Africa first took them to the Saudi Arabian Peninsula. From a man in a group of M168 haplogroup bearing tribesmen living in either Northern Africa or the Middle East came the next "marker", contained in Tim's genome; M89, which appeared about 45,000 years ago. About 90 to 95 percent of all non-Africans are descendants of this second great human migration out of Africa, and defines a large inland migration of hunters who followed expanding grasslands and plentiful game to the Middle East.
Many people of this lineage remained in the Middle East where their descendants still live, but others continued their movement and followed the grasslands through Iran to the vast steppes of Central Asia. Herds of buffalo, antelope, woolly mammoths, and other game probably enticed them to explore new grasslands.
With much of Earth's water frozen in massive ice sheets during the peak of the Wisconsinan glaciation (known as Weichselian in Europe and Devensian in the United Kingdom), the era's vast steppes stretched from eastern France to Korea. The grassland hunters of the M89 lineage traveled both east and west along this steppe "superhighway" and eventually peopled much of the continent.
A group of M89 descendants moved north from the Middle East to Anatolia and the Balkans, trading familiar grasslands for forests and high country. Though their numbers were likely small, genetic traces of their journey are still found today.
Some 40,000 years ago another direct male ancestor of Tim's, who lived in Iran or southern Central Asia, was born with a unique genetic marker that we know as M9. This marked the beginning of a new lineage that diverged from the M89 group. His descendants spent the next 30,000 years populating much of the planet.
Most residents of the Northern Hemisphere trace their roots to this unique individual, and males carry his defining marker. Nearly all North Americans and East Asians have the M9 marker, as do most Europeans and many Indians. The haplogroup defined by M9 is known as the "Eurasian Clan".
This large lineage dispersed gradually. Seasoned hunters followed the herds ever eastward along a vast belt of Eurasian steppe, into modern Iraq, Iran, Afghanistan. The Hindu Kush, Tian Shan, and Himalayas, were even more formidable barriers during the last ice age than they are today, dividing eastward migrations. These migrations through the "Pamir Knot" region would subsequently become defined by additional genetic markers.
Between 35,000 years another ancestor of Tim, Malcolm and Calder within the M9 lineage was born who bore a unique new mutation known as the M45 Marker. This man became the common ancestor of most Europeans and nearly all Native American Indians. This unique individual was part of a group of hunters who eventually broke out to the north of the mountainous Hindu Kush and onto the game-rich steppes of Kazakhstan, Uzbekistan, and southern Siberia.
The Patterson M45 lineage ancestor's who comprised the hunters were a hardy lot and managed to survive on these northern steppes even in the frigid Ice Age climate. Although big game was plentiful, these resourceful hunters had to adapt their behavior to an increasingly hostile environment. They erected animal skin shelters and sewed weather tight clothing. They also refined the flint-heads that they used on their weapons to compensate for the scarcity of obsidian and other materials.
The intelligence that followed this lineage to adapt and thrive in harsh conditions was critical to human survival in a region where no other hominids are known to have survived.
Carriers of the M45 marker moved into the Central Eurasian steppe area, some going East into Mongolia and modern China, others West into modern Russia. About 30,000 years ago another direct ancestor of Tim, Malcolm and Calder was born into the Western branch of M 45-carrying Central Asian steppe hunters. This individual and all his descendants including the Patterson's have a Y-chromosome mutation that is known as haplogroup R, which is defined by the M173 marker. Haplogroup R is characteristic of the original modern human settlers of Europe. His descendants moved across the Caucus and Ural mountains into modern Poland and Northern Germany. Some doubled back into Southern Byelorus and the Caspian Sea area.
The M173 marker bearing settlers of Western Europe, whom we know as the Cro-Magnon, immediately began to make their own dramatic mark on the continent. Famous cave paintings, like those of Lascaux and Chauvet, signal the sudden arrival of humans with artistic skill. There are no artistic precedents or precursors to their appearance.
Soon after the arrival of the M173 Cro-Magnon lineage in Europe, the Neanderthals disappeared. Recent genetic evidence proves that these hominids were not human ancestors but an evolutionary dead end. The smarter, more resourceful human descendents of M173 likely out competed the Neanderthals for scarce Ice Age resources and thus heralded their demise.
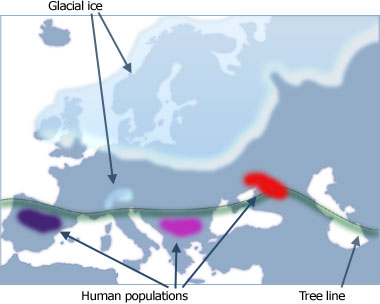
The journey of people in this lineage was strongly influenced by the large continental ice sheets that covered much of northern Europe at this time forcing them to remain in southerly refugia in Spain, Italy and the Balkans.
Map shows Palaeolithic Europe 18,000 years ago in the grip of the last ice age. Glacial ice 2km thick covers much of Northern Europe and the Alps. Sea levels are approx. 125m lower than today and the coastline differs slightly from the present day. For example, Britain and Ireland would have been connected to continental Europe (not shown on map). The air would have been on average 10-12 degrees cooler and much more arid. In between the ice and the tree line, drought-tolerant grasses and dunes would have dominated the landscape.
The westernmost members of the Cro-Magnon's gave rise to yet another direct ancestor of Tim, Malcolm and Calder as indicated by the marker M343 (R1b) which appeard in an ancestral individual who is often termed "The Patriarch", and who was born some time after 30,000 years ago. This ancestor is known as "The patriarch" because his descendants account for over 40% of all the chromosomes of Europe, and is the most common haplogroup in Europe. Soon thereafter another marker, P-25 (R1b1) appeared in an individual who is also ancestral to a large proportion of European males. The descendants of these people moved fully into Europe through modern Germany, the Low Countries, France and south into the Iberian Peninsula; a refugia where they remained until the ice began to retreat.
By 12,000 Y BP the ice has retreated and the land became much more supportive to life. Many animal species returned to inhabit the land. The three groups of humans had taken refuge for so long that their DNA had naturally picked up mutations, and consequently can be defined into different haplogroups. As they spread from these refuges, Haplogroups R1b, I and R1a propagated across Europe. The three groups of humans had taken refuge for so long that their DNA had naturally picked up mutations, and consequently can be defined into different haplogroups. As they spread from these refuges, Haplogroups R1b, I and R1a propagated across Europe.
- Haplogroup R1b is common on the western Atlantic coast as far as Scotland.
- Haplogroup I is common across central Europe and up into Scandinavia.
- Haplogroup R1a is common in eastern Europe and has also spread across into central Asia and as far as India and Pakistan.
During the last glacial maximum, R1b1 descendants produced finely knapped stone 'leaf points' which define what is known as the Solutrean culture, who were culturally distinct from the people in other European Ice Age refugia who are described more generally as Epi-Gravettian (see: Oppenheimer, Stephen. The Real Eve, pp 249-50).
The mates for R1b1 carrying males, about the time of the glacial maximum, were mtDNA haplogroups H and V. (Haplogroup V was born in the Basque area of the Pyrenees shortly after the Last Glacial Maximum. (see: Oppenheimer, Stephen. The Real Eve, p 251.)
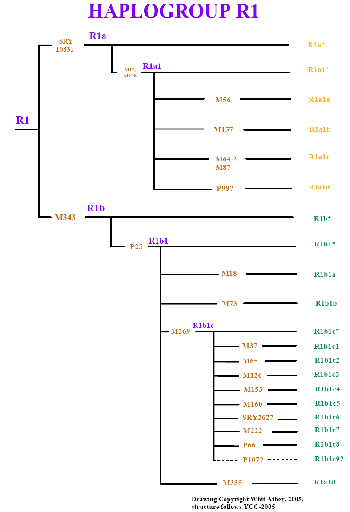
Haplogroup R1 and Subclades (click on image to see larger version.
About 12,000 years BP, following the retreat of the ice sheets, the descendants of "The Patriarch" began to agressively migrate north in large numbers, although there is evidence that they had reached the British Isles as early as 12,700 years BP. The frequency of the R1b (formerly known as Hg1 and Eu18) haplogroup changes in a cline from west (where it reaches a saturation point of almost 100% in areas of Western Ireland) to east (where it becomes uncommon in parts of Eastern Europe and virtually disappears beyond the Middle East). It is therefore often referred to as the Atlantic Modal Haplogroup (AMH).
It is believed that these people changed from hunter-gatherers to farmers in southeastern Europe about 8,000 years ago and in Britain about 4,000 years ago. As hunter-gathers became farmer's permanent settlements ended this great migration period and over time haplogroup R1b descendants settled predominately in what is known today as Spain, Portugal, France, Belgium, Denmark, England, Wales, Scotland and Ireland.
The R1b1 haplotype (a set of marker scores indicative of the haplogroup) has been difficult to interpret in that it is found at relatively high frequency in the areas where the Anglo - Saxon and later Danish "invaders" originally called home (e.g., 55% in Friesland), and even up to 30% in Norway. Thus a R1b1 haplotype makes it very challenging to interpret the origin of a family with this DNA signature. New tests are being developed which will hopefully be successful in accurately dividing R1b into subgroups that can in turn be linked to specific regions in Europe.
R1b1 haplotypes with 24 at DYS390 and 11 at DYS391 is the most populous R1b1 form and serves as the bare root of the AMH R1b1. There is historic evidence that, during the first century, BC, 'Celtic' R1b groups in the 'Germanic' part of continental, North Western Europe began to combine (and intermarry) with other groups, such as the Teutons, Slavs and Wends (who possessed R1a and I1a Y-DNA) to form homogenous yet haplotype diverse, Germanic speaking populations, to defend their territories against the Roman Empire. A large proportion of these 'Frisian-type' populations (found in present day coastal Denmark, the Netherlands and within England) who were R1b1 tended to be characterized by the genetic markers 23 at DYS390 and 11 at DYS391. This 'Frisian'-type R1b1 has about half the total popularion as the AMH R1b1 but along the Frisian coast of NW Europe the 23/11 haplotype is as abundant as the AMH form. In contrast, the proportion of 23/11haplotypes fall to very low levels in Iberia. The 'Frisian'-type R1b populations are so large and distinct that researchers speculate that Europe may have been repopulated by more than one stream of R1b coming from more than one refuge in southern Europe. Where these streams converged they began to mix to a certain extent resulting in areas dominated by AMH-type 24/11 and Frisian-type 23/11 forms but with representatives of other Rib forms present.
Recent research has shown that, later, within England, and to a lesser extent within the Scottish border areas, when the Anglo-Saxon and Danish Viking incursions took place the indigenous, ancient British 24/11 AMH-type R1b populations were largely dispersed and replaced by the invaders, whose 'Frisian-type' 23/11-R1b, R1a, and I1a Y-DNA became the dominant strains in those areas. In England, the invaders Y-DNA eventually accounted for about 67% of the newly-established 'English' male population, with the ancient British accounting for about 33%. Within the now mixed Germanic/British R1b population, of England, about 55% of the males have Germanic, 'Celtic' paternal ancestry descent, rather than ancient British. There is also very good evidence that the R1b values of 23/11 for DYS 390 and DYS391 also observered in some parts of Scotland, are also likely to have deeper Germanic-celtic paternal ancestry.
The Patterson's values for the DYS 390 and DYS391 markers are 25/11 and are distinct from both the AMH and Frisian-type markers. Based on the 12 marker results Tim's haplotype seemed most similar to a haplotype that is most abundant today in the northern part of Ireland, the west coast of Scotland. It has been variously described as the "Irish-One", "Dalriadian", or "Colla Uais" haplogroup. However, when Tim received his expanded 37 marker results this tentative placement collapsed as cluster analysis indicates that Tim's haplotype has 12 mismatches from the modal value for "Irish-One". Tim, Malcolm and Calder's haplotype is actually not very close to any R1b1 cluster with the closest match being R1b STR Cluster 39 (cluster diagram pdf courtesy of Dr. John McEwen) at 9 mismatches (MS Excel table showing quantitative relationship between Tim's haplotype and other R1b STR clusters courtesy of Dr. David Wilson).The problem with intepreting many R1b haplotypes is that members of this haplogroup have diverged from the original founder under conditions where there has been almost continuous population expansion (along with the odd pruning with plagues and such). In R1b there are branches which have branched over time, and which have in turn branched again and again in almost all possible variants. This mode of almost continuous expansion within R1b makes it very difficult to calculate the exact branching structure, a good reason to consider clusters instead.
At this stage a few possible scenarios for the origin of the Patterson's haplotype can be speculated upon:1) taking a basic subset of Tim's haplotype (9 markers) and entering it into YHRD provided no identical matches. The largest group 1 step distant had 9 hits scattered across western Europe with a hint that the haplotype is centered in western Europe. At two steps distant most matches were in western Europe (mostly Iberia and the British Isles) with a slightly higher precentage in Ireland -- basically the distirubiton of R1b as a whole.
2) An assessment of the surnames within the closest cluster to Tim's haplotype, R1bSTR39, do not turn up many names that look very Irish ordal Riadic Scots. However, care must be taken because the exisitng databases are biased.
3) Looking at the closest 37 marker matches in FTDNA reveals several results with mismatches 8-10 genetic distant away (i.e. not all that close), many of which were from Ireland or Scotland. This linkage provides some suggestion that this variant may have been based at one time in Northern Ireland.
The best guess of geneticist John McEwen is that the haplotype is most likely of English/Anglosaxon origin assuming that the Patterson haplotype actually belongs to the closest cluster R1bSTR39.
Dr. McEwen's tentative conclusion is that:
The Patterson's have a rarer haplotype that does not fit well with existing clusters.
Tim's closest matches appear to come from Ireland and Scotland.
That is about all that can concluded about the Patterson haplotype at this at this stage. As more data becomes available rarer variants such and that of the Patterson's will have more members and an idea of likely origins will be clearer. The next stage will be to have SNP testing done to determine better how the Patterson's fit into the bigger R1b picture.
Definitions
Allele: Alternative form of a genetic locus; a single allele for each locus is inherited separately from each parent (e.g., at a locus for eye color the allele might result in blue or brown eyes).
DYS : D = DNA, Y = Y chromosome, S = a unique DNA segment. This nomenclature is controlled by the HUGO Gene Nomenclature Committee, with the assignment of new DYS numbers. This guideline determines each part of the symbol for naming arbitrary DNA fragments and loci. See section Appendix App 1.1 DNA Segments located at:
http://www.gene.ucl.ac.uk/nomenclature/guidelines.html#1.4Locus (pl. loci): The position on a chromosome of a gene or other chromosome marker; also, the DNA at that position. The use of locus is sometimes restricted to mean regions of DNA that are expressed. The specific physical location of a gene on a chromosome. From the Latin for 'place'. A stretch of DNA at a particular place on a particular chromosome — often used for a 'gene' in the broad sense, meaning a stretch of DNA being analyzed for variability (e.g., a microsatellite locus).
Marker: An identifiable physical location on a chromosome (e.g., restriction enzyme cutting site, gene) whose inheritance can be monitored. Markers can be expressed regions of DNA (genes) or some segment of DNA with no known coding function but whose pattern of inheritance can be determined. A gene of known location on a chromosome and phenotype that is used as a point of reference in the mapping of other loci
Return to DNA Test Results Index
Link to table where Y chromosome marker results for all participants are summarized together.