Return to DNA Test Results Index
Link to table where Y chromosome marker results for all participants are summarized together
Y Chromosome Test Results For
Speedy/Moffitt Descendant D. Kevin Locke
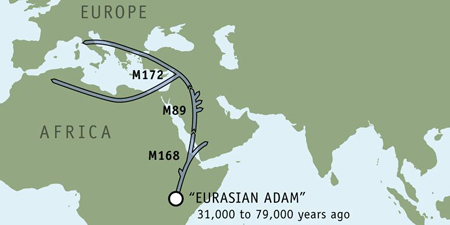
Kevin's Y chromosome results identify him as a member of haplogroup J2, a lineage defined by a genetic marker called M172. This haplogroup is the final destination of a genetic journey that began some 60,000 years ago with an ancient Y chromosome marker called M168. J2 haplotypes are very rare in people originating from the United Kingdom.
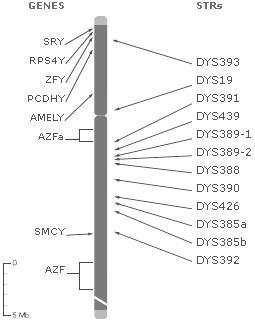
The test results below are from the laboratory analysis of Kevin's Y-chromosome. His DNA was analyzed for Short Tandem Repeats (STRs), which are repeating segments of the genome that have a high mutation rate. See definitions of Locus, DYS#, and Alleles at the bottom of this page.
Locus |
DYS# |
Alleles |
|
1 |
393 |
12 |
|
2 |
390 |
24 |
|
3 |
19 (also 394) |
15 |
|
4 |
391 |
9 |
|
5 |
385a |
11 |
|
6 |
385b |
17 |
|
7 |
426 |
11 |
|
8 |
388 |
16 |
|
9 |
439 |
11 |
|
10 |
389-1 |
14 |
|
11 |
392 |
11 |
|
12 |
389-2 |
29 |
|
Kevin who lives in Texas is a Speedy/Moffitt descendant via Mary Ann Moffitt who was the daughter of Andrew Moffitt & Margaret "Jane" Piercy. Mary Ann's husband was William A. Speedy, the son of Thomas Speedy and Isabella Patterson. Kevin's Y chromosome values comes to him via his direct male ancestor Capt. John Locke who founded Locke's Neck, New Hampshire in the mid 1600's.
The location on the Y chromosome of each of the tested markers is depicted in the image above, with the number of repeats for each of your STRs presented to the right of the marker. For example, DYS19 is a repeat of TAGA, so if Kevin's DNA repeated that sequence 12 times at that location, it would appear: DYS19 12. Studying the combination of these STR lengths in a persons Y Chromosome allows researchers to place individuals in a haplogroup, which reveals the complex migratory journeys of ancestors.
The very widely dispersed M168 marker can be traced to a single individual--"Eurasian Adam." This African man, who lived some 31,000 to 79,000 years ago, is the common ancestor of every non-African person living today. His descendants migrated out of Africa and became the only lineage to survive away from humanity's home continent.
Population growth during the Upper Paleolithic era may have spurred the M168 lineage to seek new hunting grounds for the plains animals crucial to their survival. A period of moist and favorable climate had expanded the ranges of such animals at this time, so these nomadic peoples may have simply followed their food source.
Improved tools and rudimentary art appeared during this same epoch, suggesting significant mental and behavioral changes. These shifts may have been spurred by a genetic mutation that gave "Eurasian Adam's" descendants a cognitive advantage over other contemporary, but now extinct, human lineages.
Some 90 to 95 percent of all non-Africans are descendants of the second great human migration out of Africa, which is defined by the marker M89.
M89 first appeared 45,000 years ago in Northern Africa or the Middle East. It arose on the original lineage (M168) of "Eurasian Adam," and defines a large inland migration of hunters who followed expanding grasslands and plentiful game to the Middle East.
Many people of this lineage remained in the Middle East, but others continued their movement and followed the grasslands through Iran to the vast steppes of Central Asia. Herds of buffalo, antelope, woolly mammoths, and other game probably enticed them to explore new grasslands.
With much of Earth's water frozen in massive ice sheets, the era's vast steppes stretched from eastern France to Korea. The grassland hunters of the M89 lineage traveled both east and west along this steppe "superhighway" and eventually peopled much of the continent.
A group of M89 descendants moved north from the Middle East to Anatolia and the Balkans, trading familiar grasslands for forests and high country. Though their numbers were likely small, genetic traces of their journey are still found today.
Haplogroup J2, defined by marker M172 originated in the Fertile Crescent and marks a major milestone of the human journey. Some 10,000 to 15,000 years ago the members of this group became the first farmers during a period known as the Neolithic Revolution. With the success of agriculture, they later pioneered the rise of modern, sedentary communities and cities.
M172 defines a major subset of this haplogroup which arose from the M89 lineage. It is found today in North Africa, the Middle East, and southern Europe. In southern Italy it occurs at frequencies of 20 percent, and in southern Spain ten percent of the population carries this marker.
This lineage's early farming successes spawned population booms and encouraged migration throughout much of the Mediterranean world. Later migrations carried M172 through the river valleys of Central Asia and into northern India.
Descendants of M172 have left a physical footprint that matches their genetic journey. Sites such as Jericho (Tell el-Sultan) are among the world's oldest settled communities, and allow archaeologists to trace many millennia of growth and development.
The J2 haplogroup came to England either through middle Eastern Roman soldiers who were stationed on the island (most common explanation), through Sephardic traders (not many of those), or through the migration to the island of gypsies in the 16th century (only being thought of as a possible source very recently).
Definitions
Allele: Alternative form of a genetic locus; a single allele for each locus is inherited separately from each parent (e.g., at a locus for eye color the allele might result in blue or brown eyes).
DYS : D = DNA, Y = Y chromosome, S = a unique DNA segment. This nomenclature is controlled by the HUGO Gene Nomenclature Committee, with the assignment of new DYS numbers. This guideline determines each part of the symbol for naming arbitrary DNA fragments and loci. See section Appendix App 1.1 DNA Segments located at:
http://www.gene.ucl.ac.uk/nomenclature/guidelines.html#1.4Locus (pl. loci): The position on a chromosome of a gene or other chromosome marker; also, the DNA at that position. The use of locus is sometimes restricted to mean regions of DNA that are expressed. The specific physical location of a gene on a chromosome. From the Latin for 'place'. A stretch of DNA at a particular place on a particular chromosome — often used for a 'gene' in the broad sense, meaning a stretch of DNA being analyzed for variability (e.g., a microsatellite locus).
Marker: An identifiable physical location on a chromosome (e.g., restriction enzyme cutting site, gene) whose inheritance can be monitored. Markers can be expressed regions of DNA (genes) or some segment of DNA with no known coding function but whose pattern of inheritance can be determined. A gene of known location on a chromosome and phenotype that is used as a point of reference in the mapping of other loci.
Return to DNA Test Results Index
Link to table where Y chromosome marker results for all participants are summarized together