Haplogroup J Distribution
Return to mtDNA Test Results Summary Page
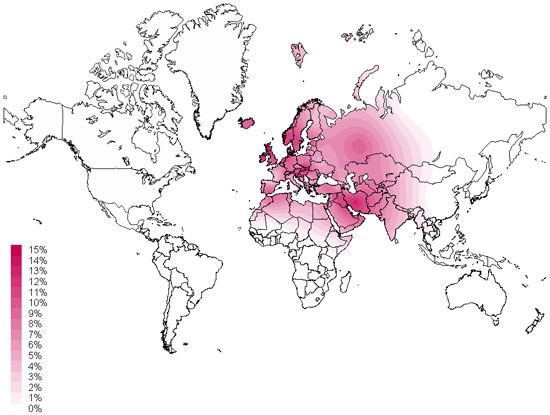
The map above produced by the Sorenson Molecular Genealogy Foundation map shows the geographic distribution of haplogroup J as measured in various geographically and ethnically defined populations from around the world. The color values displayed on the global map correspond to the percentage of individuals at that location who belong to this Haplogroup.
Origins
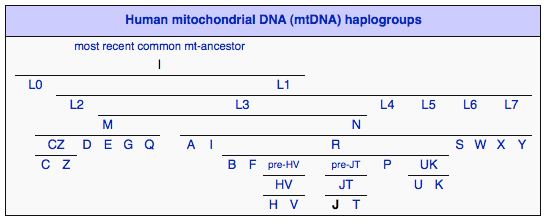
Haplogroup J is a human mitochondrial DNA (mtDNA) haplogroup. Haplogroup J derives from the haplogroup JT, which also gave rise to Haplogroup T. In his popular book The Seven Daughters of Eve, Bryan Sykes named the originator of this mtDNA haplogroup "Jasmine". Around 45,000 years before present, a mutation took place in the DNA of a woman (Jasmine) who lived in the Caucasus region. Further mutations took place in the J line which can be identified as J1a1 (27,000 yrs ago), J2a (19,000 yrs ago), J2b2 (16,000 years ago), J2b3 (5,800 yrs ago), etc.
Haplogroup J is a sister to haplogroup T because both variations diverged from a common ancestor that was living in the Near East during the Paleolithic era. Indeed the high regional diversity of haplogroup J in the Near East supports the idea of Levantine origins - present-day areas of Israel, Jordan, Lebanon, Palestine, Syria and the Sinai Peninsula. Later the spread of some J variants into Caucasus, Europe and North Africa is associated with the Neolithic period, when humans were developing the skills of farming and herding, approximately 10,000 years ago. Haplogroup J (along with ‘T’) has thus been associated with the spread of farming and herding in Europe during the Neolithic Era (8,000-10,000 yrs ago). All other West Eurasian-origin groups (H, V, U, K, W, I, X) were previously given to hunting and gathering. Taking into account its wide regional diversity, it is possible haplogroup J was involved in several expansion events other than the one associated with the Neolithic period.
Two main subclades within Haplogroup J exist; J1 and J2. The J1 subclade accounts for nearly 70% of the total J population; however, the J2 subclade is 10,000 years older than J1. Thus, if the late arrival of J in Europe is associated with the expansion of Neolithic farmers from the Near East (see above), it is more plausible that its sub-branch J1 was partly involved in this process.
mtDNA Haplogroup J Distribution
Mitochondrial haplogroup J is typically found in Western Eurasian populations. The distribution of haplogroup J stretches from the Near East (12% of the population) to the Caucasus region (8% of the population), into Europe (10% of the population) and North Africa (6% of the population). Haplogroup J reaches its highest frequencies in Arabia, particularly in Bedouin and Yemeni groups where the variation is found in 25% of these populations.
The J1 subclade is spread throughout the European continent at the following frequencies; Britain (10% of the J population), Germany (8%), Austria (5%), Switzerland (5%), France (3%) and Italy (3%).
The occurrence of J2 is more localized along the Mediterranean, particularly the Balkan Peninsula; approximately 50% of the Greek population carries the J2 variation. However, J2 populations are particularly diverse, ranging from the Mansi and other Ob River populations in Western Siberia (10% of the J2 population) to populations of Northern Iran (5% of the J2 population).
mtDNA Haplogroup J Distribution within Europe
Within Europe, >2% frequency distribution of mtDNA J is as follows [2]:
J* = Ireland - 12%, England-Wales - 11%, Scotland - 9%, Orkney - 8%, Germany - 7%, Russia (European) - 7%, Iceland - 7%, Austria-Switzerland - 5%, Finland-Estonia - 5%, Spain-Portugal - 4%, France-Italy - 3%
J1a = Austria-Switzerland - 3%
J1b1 = Scotland - 4%
J2 = France-Italy - 2%
J2a = Homogenously spread in Europe. Absent in the nations around the Caucasus. Not known to be found elsewhere.
J2b1 = Virtually absent in Europe. Found in diverse forms in the Near East.
References
1. Caramelli D, Lalueza-Fox C. et al, Genetic analysis of the skeletal remains attributed to Francesco Petrarca. Forensic Sci Int. (Feb. 2007)
2. Palanichamy MG, Sun C, et al. Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet. (Dec. 2004)
3. Richards M, Macaulay V, et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. (Nov. 2000)